February 10, 2020 - Continuing to Improve Abstract and Seeing Replicability of Image Segmentation
Lab Work
Today, I continued to look through the abstract for the Undegraduate Research Symposium and edited it according to the comments that Laura gave me. Dr. Roberts and Laura approve of it so I will include it in my application and send it in by Wednesday!
Today, I also tested the script that I have been using on another image today to analyze its replicability, and overall, it did a pretty good job once again (see resulting images below):
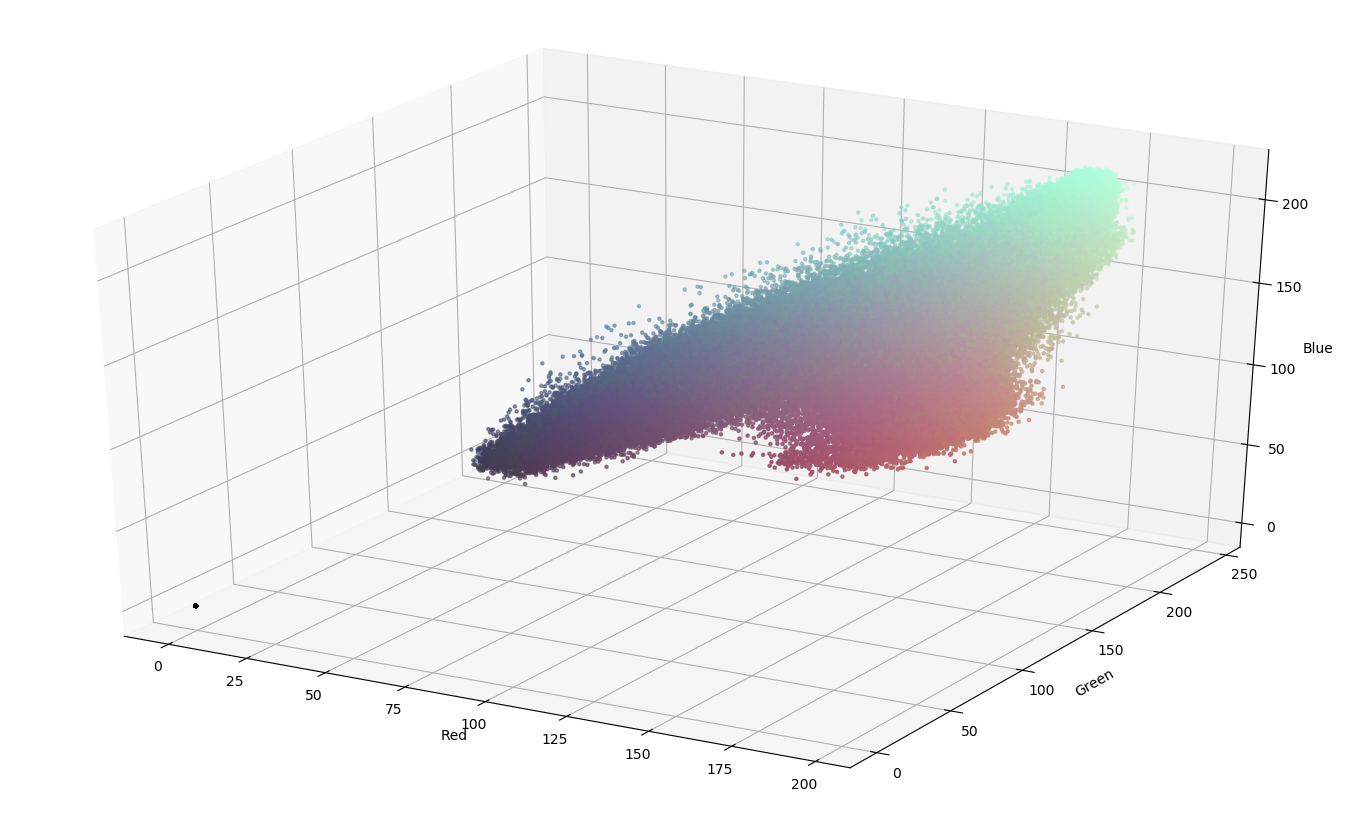
Figuring out color ranges on color scatter plot:
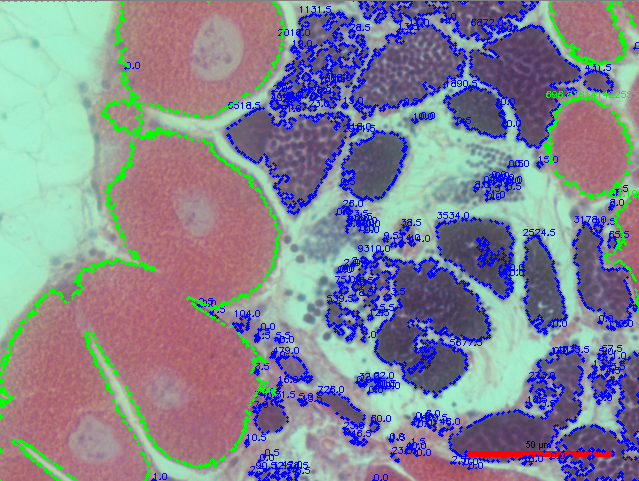
Resulting Threshold Image:
Another thing that I figured out along the way was how to make the resulting image in RGB instead of BGR, which is great! In addition, I have created a protocol for how I am currently doing the image segmentation here, and I will improve it over time as I improve the methodology itself.
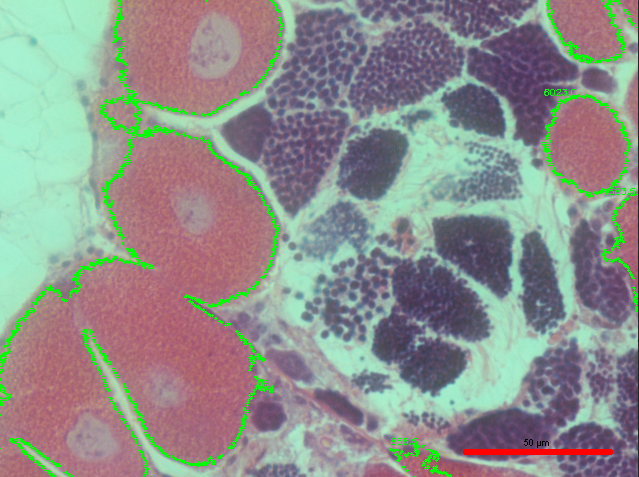
But, although the script did a pretty good job, there is still definitely room for improvement. For example, not all of the pink female gonad was marked in the image. So, in order to fix this, I reduced the size threshold, and this is the resulting output:
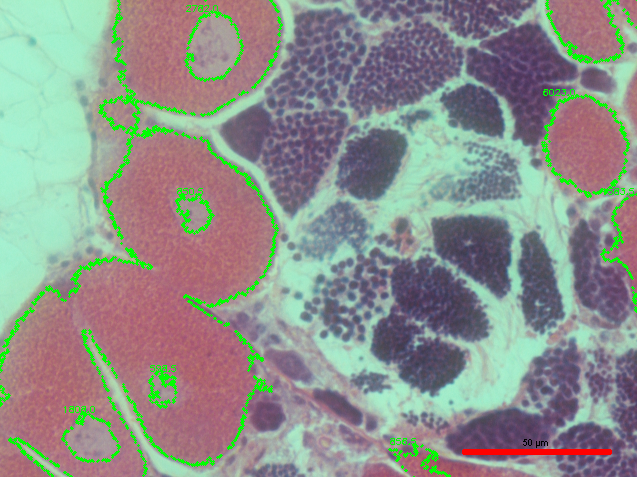
The size threshold seemed to help a little, but there is still some missing gonad that I would like for the script to recognize.
An additional problem that I noticed is that there are a lot less objects being identified as eggs than there are actual eggs present in the image:
Once I looked into it some more, I realized that the reason for this is because there is one huge group of eggs that are clustered together, and this group is being counted as 1 egg because they are all connected to each other. This would still be a viable way to figure out what sex the organism is (male only, female only, hermaphrodite predominantly male, hermaphrodite predominantly female, etc…), but it is not a viable way to measure each individual egg in the image. Thus, I might have to add circularity as a factor to measure each egg or utilize another function such as a watershed algorithm. The links below may help me figure out how to use the watershed algorithm in OpenCV:
Something that might be useful in the future - if want to find nucleus size, I could use something like (I found this out through this link):
- RETR_TREE
instead of
- RETR_EXTERNAL
to produce images such as this:
One thing that I realized while rerunning the script on a different image is that setting different color ranges for male and female gonads each time may become a hassle if I have to do that for every single image; so I may want to look into if there are patterns of coloration for male and female gonads throughout the images.
Next Steps
- See if watershed algorithm method may be a good way to separate and measure individual eggs in image
- Find a possible pattern of coloration in order to make it easier to quantify gonadal sex and stage with less bias